December 2021 ... Matthew Lee presents his and Bodi's manuscript, Characterizing the Behavior of Mutated Proteins with EMCAP: the Energy Minimization Curve Analysis Pipeline, at CSBW (virtual) at IEEE BIBM.
Alistair Turcan presents our manuscript, Assessing the Effects of Amino Acid Insertion and Deletion Mutations, at CSBW (virtual) at BIBM.
The CGRAP server is launched, and published (CGRAP: A Web Server for Coarse-Grained Rigidity Analysis of Proteins) in Symmetry. Kudos to everyone who contributed to the long-term effort ... Alistair, Anna, Dylan, Lorraine, and Lauren.
October 2021 ... collaboration work with Dr. Amacher's lab is published, Domain Analysis and Motif Matcher (DAMM): A Program to Predict Selectivity Determinants in Monosiga brevicollis PDZ Domains Using Human PDZ Data, in Molecules
September 2020 ... Cecilia Kalthoff and Gideon Wolfe Present their teams' spring 2020 bioinformatics projects, accepteed as manuscripts to CSBW, at ACM-BCB 2020 (virtual): Assessing Drug Resistance Due to Mutations via Energy Minimization Profiles, and Impactful Mutations in Mpro of the SARS-CoV-2 Proteome
September 2019 ... Sam Herr presented his paper, PETRA: Protein-ligand Complex Engineering Through Rigidity Analysis at CSBW at ACM BCB in Niagara Falls, NY.
July 2019 ... Tess Thackray, Bodi Van Roy, and Dylan Carpenter (not pictured) presented two posters at the 2019 Protein Sympoium in Seattle. Tess and Bodi presented their on-going work on Protein resilience, while Dylan presented his on-going masters work in generating mutants of protein ligand complexes and their assessment.
July 2018 ... Nicholas Majeske presented his manuscrtip, Elucidating Which Pairwise Mutations Affect Protein Stability: An Exhaustive Big Data Approach, at COMPSAC 2018, in Tokyo. Nicholas is now a PhD student at Indiana University Bloomington.
March 2018 ... Hunter Read presented his paper, cMutant : A Web Server and Compute Pipeline for Exploring the Effects of Amino Acid Substitutions via Rigidity Mutation Maps, and Richard Olney presented Protein Mutation Stability Ternary Classification using Neural Networks and Rigidity Analysis, at the 10th International Conference on Bioinformatics and Computational Biology (BICOB) in Las Vegas. Great job Hunter and Richard, and to the other WWU student authors of those two publications.
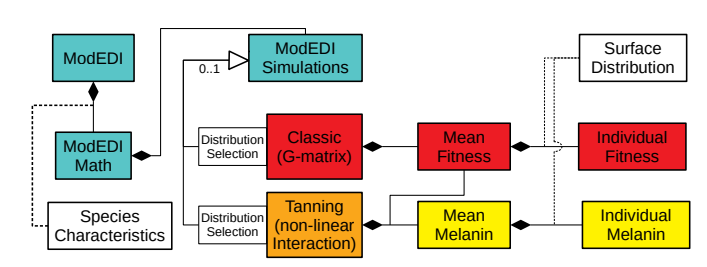
March 2018 ... Elizabeth Brooks has been accepted to the PhD program in biological sciences at Notre Dame. She has been working in our lab with Graham Roberts, co-supervised by Ali Scoville of CWU, on a software suite of programs for exploring the effects of nonlinear developmental interactions. She and Graham presented their paper, ModEDI: An Extendable Software Architecture for Examining the Effects of Nonlinear Developmental Interactions on Evolutionary Trajectories, at the IEEE ICCABS conference in October 2017. Congrats Elizabeth!
December 2017 ... Kyle Daling presented his work, Contrained Sequence Alignment, to fulfill and satisfy the requirements for the Honors Program at WWU. More information is at WWU Honors, and Honors Presentation pages.
October 2017 ... graduate student Elizabeth Brooks and undergraduate student Graham Roberts presented their paper, ModEDI: An Extendable Software Architecture for Examining the Effects of Nonlinear Developmental Interactions on Evolutionary Trajectories, at the IEEE ICCABS 2017 conference in Orlando, Florida. Work is on-going, and we hope to submit a journal manuscript in the next year. Good job Elizabeth and Graham.
October 2017 ... Stephanie Mason won first place at the ACM undergraduate research competition (SRC) at the Grace Hopper Conference for her bioinformatics work currently underway in our lab. She's now a grand finals candidate. Well done Stephanie! And congrats to Ted, too, who also was honored. Stephanie's manuscript and a news article are available at the ACM Digital Library, and Western Today.