Lecture 17 - Machine Learning: Feature Extraction; Vectors and Distances¶
Announcements:¶
Goals:¶
- Understand the purpose of feature extraction, and the meaning of feature vector
- Know the meaning and purpose (but not yet the mechanism) of clustering and dimensionality reduction
- Know how and when to use various distance metrics to compare feature vectors:
- $L^p$ distances, including $L^2, L^1, L^0$, and $L^\infty$
- Hamming distance
- Cosine similarity
- Gain a basic understanding of the curse of dimensionality as pertains to vector distances
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
ML For Data Science: Unsupervised Learning¶
As data scientists, we are often looking to discover trends, patterns, or underlying structure in data. In contrast with prediction problems, we often don't have a particular quantity we're interested in predicting, but rather we want to gain insights from data.
Here we're going to demo a couple unsupervised techniques - clustering and dimensionality reduction.
Penguins Pairplot¶
We're going to work with the Palmer Penguins dataset.
penguins = sns.load_dataset("penguins").dropna()
penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
333 rows × 7 columns
Feature Extraction, Version 0.0¶
Machine learning methods pretty much universally operate on vectors-1D arrays of numbers. Often your first job as an ML practitioner is to turn whatever you have (e.g., penguins) into vectors.
Someone's done the hard work of measuring a bunch of quantities for each penguin; for now, we're just going to take the for numerical columns and treat them as a length-4 ("four dimensional", or "4D") feature vector. Later we may need to get fancier than this.
# create the dataset - get the numerical columns
df = penguins.copy(deep=True)
numerical_features = [
'bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g'
]
X = df[numerical_features]
X
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| 0 | 39.1 | 18.7 | 181.0 | 3750.0 |
| 1 | 39.5 | 17.4 | 186.0 | 3800.0 |
| 2 | 40.3 | 18.0 | 195.0 | 3250.0 |
| 4 | 36.7 | 19.3 | 193.0 | 3450.0 |
| 5 | 39.3 | 20.6 | 190.0 | 3650.0 |
| ... | ... | ... | ... | ... |
| 338 | 47.2 | 13.7 | 214.0 | 4925.0 |
| 340 | 46.8 | 14.3 | 215.0 | 4850.0 |
| 341 | 50.4 | 15.7 | 222.0 | 5750.0 |
| 342 | 45.2 | 14.8 | 212.0 | 5200.0 |
| 343 | 49.9 | 16.1 | 213.0 | 5400.0 |
333 rows × 4 columns
We're secretly interested in the Species column, which the clever scientists have already figured out, but for hte sake of argument we're pretending that we don't know this. As such, this is our y, a quantity of interest that - in our unsupervised context - we might want to discover.
y = df['species']
y
0 Adelie
1 Adelie
2 Adelie
4 Adelie
5 Adelie
...
338 Gentoo
340 Gentoo
341 Gentoo
342 Gentoo
343 Gentoo
Name: species, Length: 333, dtype: object
Let's look at a pairplot of the numerical columns:
sns.pairplot(data=penguins);
Looking at all those scatterplots, it seems like there's some pattern here - the penguins seem to be grouped into 2 or 3 clusters. Hmm...
We're going to apply the k-means clustering to put the penguins into three groups based on their mutual similarity. I've hidden away the inner workings inside cluster_demo_fit and cluster_demo_vis - for now, the goal is simply to see the result of a clustering algorithm, not understand how it works (we'll get there soon!).
# ignore this code for now!
from sklearn.cluster import KMeans
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import adjusted_rand_score
def cluster_demo_fit(X):
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
kmeans = KMeans(n_clusters=3, random_state=42)
y_pred = kmeans.fit_predict(X_scaled)
return y_pred
def cluster_demo_vis(df, x_col, y_col):
fig, axes = plt.subplots(1, 2, figsize=(8, 4))
# Left Plot: True Species Labels
sns.scatterplot( data=df, x=x_col, y=y_col, hue='species', palette='viridis', ax=axes[0], s=20, alpha=0.8)
axes[0].set_title('Ground Truth (Actual Species)')
axes[0].legend(title='Species')
# Right Plot: Predicted Cluster Labels
sns.scatterplot(data=df, x=x_col, y=y_col, hue='cluster', palette='Set1', ax=axes[1], s=20, alpha=0.8, legend='full')
axes[1].set_title(f'K-Means Clusters (K=3)')
axes[1].legend(title='Cluster ID')
plt.show()
# cluster_demo_fit returns a column with the cluster number for each datapoint
df['cluster'] = cluster_demo_fit(X)
df
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | cluster | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male | 0 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female | 0 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female | 0 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female | 0 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female | 1 |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female | 1 |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male | 1 |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female | 1 |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male | 1 |
333 rows × 8 columns
# choose 2 axes to visualize along
x_col = 'flipper_length_mm'
y_col = 'bill_length_mm'
cluster_demo_vis(df, x_col, y_col)
# Confusion matrix showing cluster assignment vs species
comparison_table = pd.crosstab(df['species'], df['cluster'])
sns.heatmap(comparison_table, annot=True, fmt='g')
<Axes: xlabel='cluster', ylabel='species'>
# again, ignore the code!
from sklearn.random_projection import GaussianRandomProjection
df = penguins.copy(deep=True)
def dim_reduction_demo(X):
scaler = StandardScaler()
n_components = 2
rp = GaussianRandomProjection(n_components=n_components)
X_proj = rp.fit_transform(X_scaled)
return X_proj
df[["D1", "D2"]] = dim_reduction_demo(X)
plt.figure(figsize=(6, 4))
sns.scatterplot(x='D1', y='D2', hue='species', data=df, palette='viridis', s=20)
plt.title('Dimensionality Reduction (4D to 2D) of Palmer Penguins Dataset')
plt.xlabel(f'Reduced Dimemsion 1')
plt.ylabel(f'Reduced Dimemsion 2')
plt.grid(True)
plt.legend(title='Species')
plt.show()
Distance Metrics¶
All of the above methods require the fundamental ability to compare datapoints, in particular to answer the question: how similar or different are they?
$L^p$ Distances¶
A common family of distance metrics is the $L^p$ distance:
$$d_p(a, b) = \sqrt[p]{\sum_{i=1}^d |a_i - b_i|^p}$$
When $p = 2$, this is the Euclidean distance we're all used to, based on the Pythagorean theorem; in 2D, it reduces to: $$\sqrt{(b_x - a_x)^2 + (b_y - a_y^2)}$$
Different values of $p$ give different behavior:
- For smaller $p$, we care less about how different the per-dimension differences are from each other.
- For larger $p$, we care more about how different the per-dimension differences are from each other.
A few examples of the "unit circle" under different $L^p$ distances:
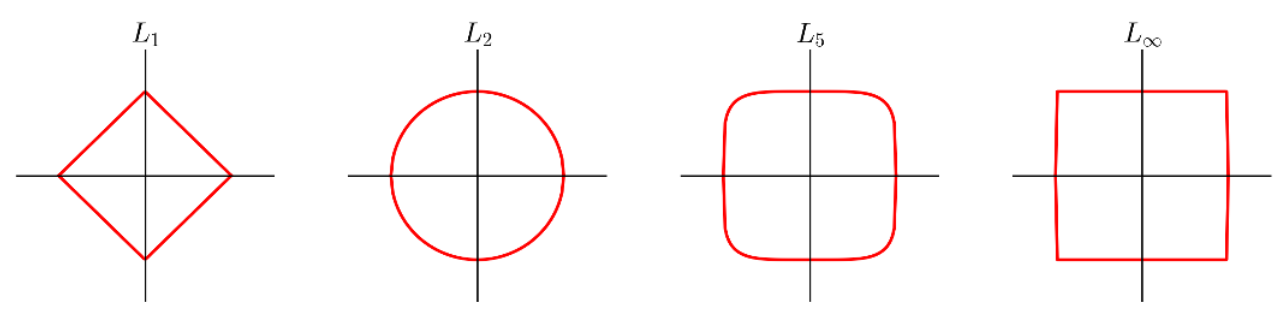
$L_1$ and $L_2$ are by far the most common choices here.
Exercise: rank the penguins by similarity to Penguin 0, based on L1, L2, distance between flipper + bill length vectors.
penguins.iloc[0:1,:]
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
Start by writing a function to compute $L^p$ distance between two vectors:
def L(p, a, b):
""" Compute the L^p distance between vectors a and b
Pre: p > 0 and a, b are d-dimensional 1d arrays """
return np.sum(np.abs(a - b) ** p) ** (1/p)
L(2, np.array([1, 1]), np.array([2,2]))
# penguin_0 = penguins.iloc[0][numerical_features]
np.float64(1.4142135623730951)
Now create a new column in penguins with the $L^2$ distance to penguin 0:
penguins["L1"] = penguins[numerical_features].apply(lambda b: L(1, penguin_0, b[numerical_features]), axis=1)
#sns.pairplot(data=penguins)
Complete a similar process for L^1 distance.
More Distance Metrics¶
Hamming Distance¶
For vectors of categorical values, Hamming distance is the number of dimensions in which two vectors differ: $$d(a, b) = \sum_i \mathbb{1}(a_i \ne b_i)$$ where $\mathbb{1}(\cdot)$ is an indicator function that has value 1 if its argument is true and 0 otherwise.
Cosine Similarity¶
A similarity (not distance) metric that considers only vector direction, not magnitude:
$$ sim(a, b) = \cos \theta = \frac{a^Tb}{\sqrt{(a^Ta)(b^Tb)}}$$