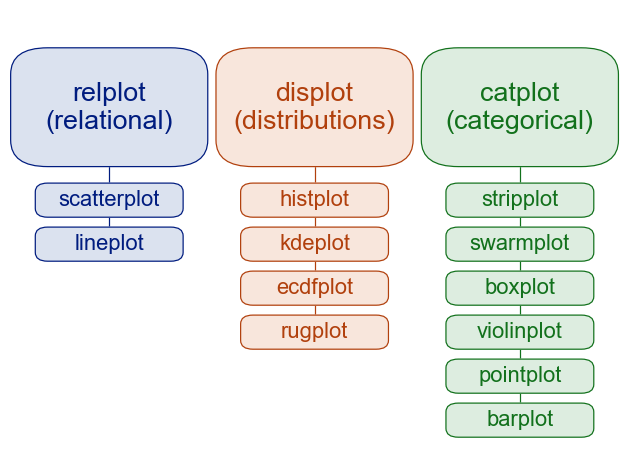
colors = {"Adelie": "red", "Gentoo": "green", "Chinstrap": "blue"}
size = lambda x: 10 if x > 40 else 1
plt.scatter("body_mass_g", "flipper_length_mm", data=penguins,
c=penguins["species"].map(colors),
s=(penguins["bill_length_mm"])/4)
plt.legend()
plt.xlabel("Body Mass (g)")
plt.ylabel("Flipper Length (mm)")
Text(0, 0.5, 'Flipper Length (mm)')